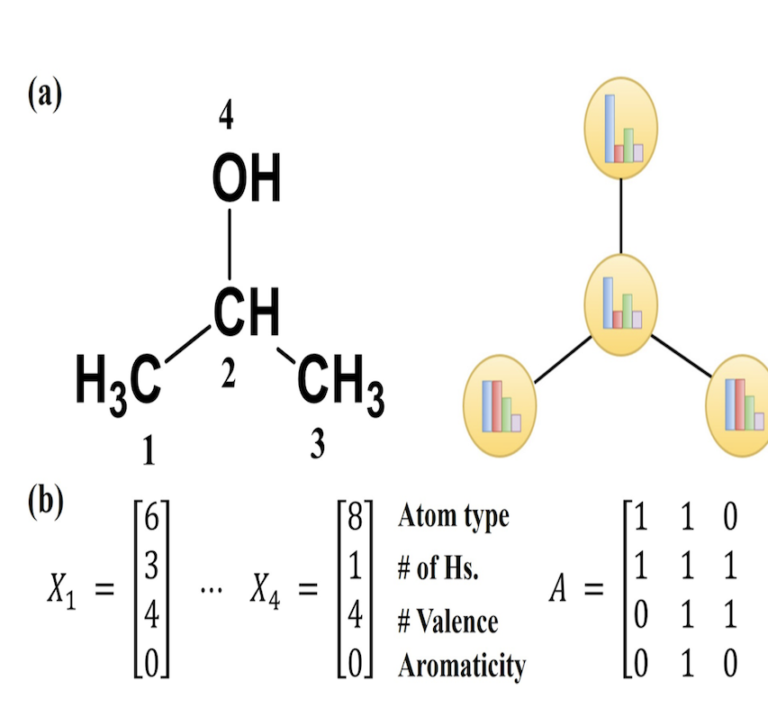
Transcriptomics CHO Alignment Pipeline
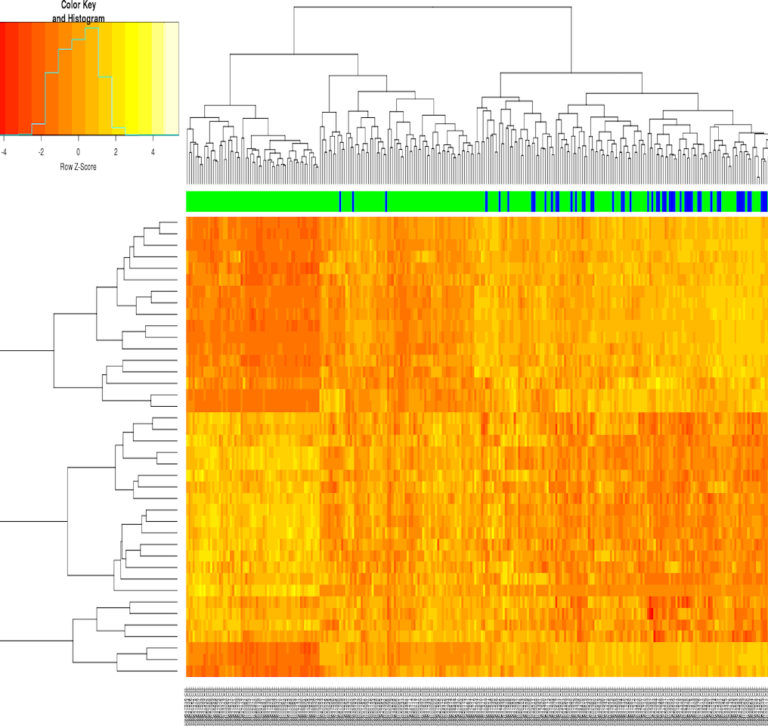
Summary Using fastqc, HISAT2, featureCounts, samtools, and Trimmomatic, an alignment pipeline was made for RNAseq data to the Chinese hamster ovary (CHO) genome. The annotation files were taken from NCBI. The workflow until the featureCounts step show’s an older annotation file. Note that the workflow is identical for the updated annotation file, but the updated …