Source Code: GitHub
Summary (Not Completed!)
Exploration of a dataset related to readmission of diabetes patients based on attributes the hospitals have collected. The goal of this workflow was to see if there is a way to successfully create a classification model that could differentiate between no admission, readmission before 30 days, and readmission after 30 days. The motivation behind a hospital knowing this has to do with financial burdens that readmission can put on patients. With this information, perhaps hospitals could decide to prescribe more effective treatments. The scope of the use of this information is up to the end user, but there are various ways you could use the readmission rates to more effectively run a hospital.
Data Background
The data for this project comes from 130 US hospitals ranging from 1999-2008. The full background can be found here.
Technical Background
This project utilized the following algorithms to assess their performance on predicting the diabetes dataset.
- Gradient Boosting (Xgboost)
- Random Forest
- Nueral Network (MLP)
The training data involved X features and 3 classes for the target. Before digging into the actual model building, some exploratory data analysis was done using PCA to see if the classes were linearly separable.
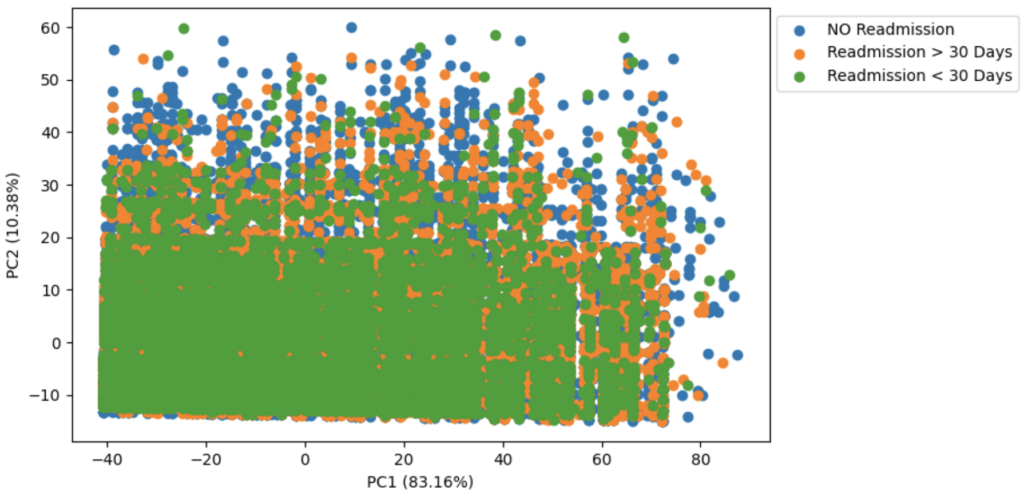

Evidently there is almost no separation between these classes using Principal Component Analysis. Given autoencoder’s performed better than the principal component analysis in the RNA Transcriptomics Analysis project, it was worth seeing if the autoencoder structure could find more meaningful separation planes. The autoencoder has the following notable features
- Latent Dimension of XX
- Embedding input_dim of