Source Code: GitHub
Competition Statement
The goal of this competition is to predict if a person has any of three medical conditions. You are being asked to predict if the person has one or more of any of the three medical conditions (Class 1), or none of the three medical conditions (Class 0). You will create a model trained on measurements of health characteristics.
To determine if someone has these medical conditions requires a long and intrusive process to collect information from patients. With predictive models, we can shorten this process and keep patient details private by collecting key characteristics relative to the conditions, then encoding these characteristics.
Your work will help researchers discover the relationship between measurements of certain characteristics and potential patient conditions.
Section 1: Final Approach
Project General Approach
The first step in analyzing this data was to do some preprocessing. The following were the two preprocessing steps that were used. These techniques, or some similar alternative, are standard practice when doing data analysis for machine learning models.
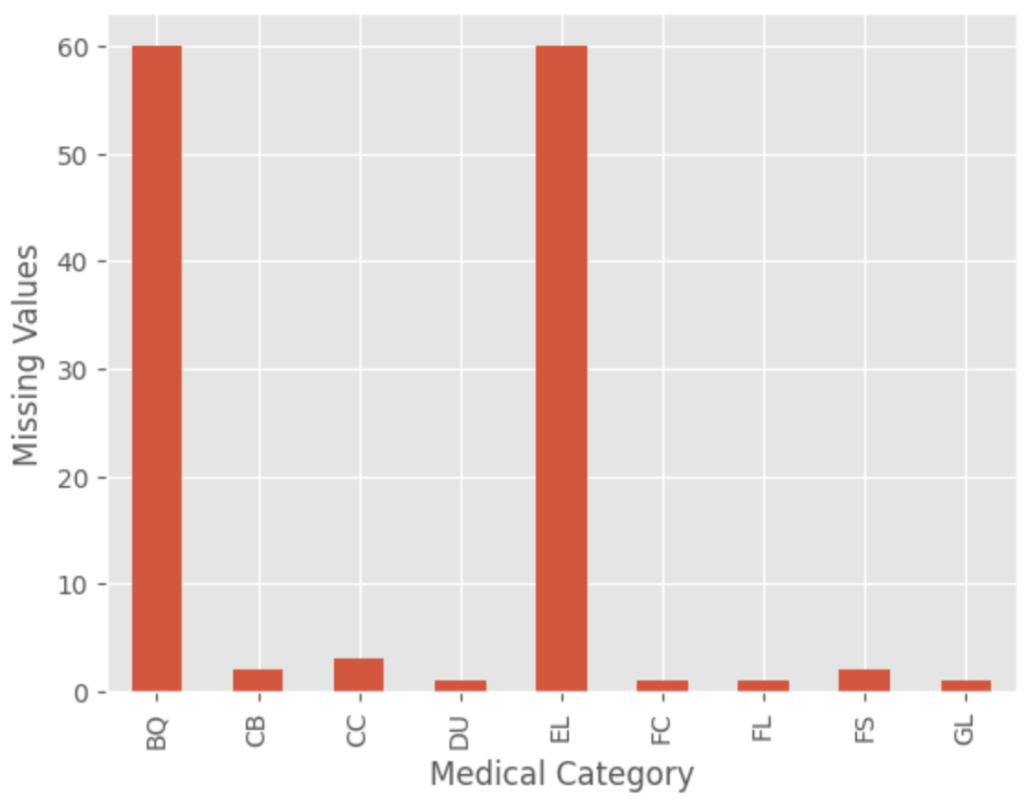
- Check the data for missing values and impute the values using a random forest imputer
- Understand how much of the data is categorical or continuous
- Normalize the data
The next step after preprocessing was to decide if I wanted to embed the normalized data in any way. One great characteristic of embedding the data is that at times it can be more robust to covariate shift that occurs from having different types of test data.
I didn’t end up doing any embeddings for my features, but after the competition closed, I saw the second-place winner chose to use UMAP embeddings. To this, I revert to the No Free Lunch Theorem for machine learning, which in short states that no one embedding or model will always produce the best results. It will always depend, which is why it’s wise to create multiple models and constantly observe their performance. Anyway, back to the competition workflow
After deciding that I would not embed my data, I had to consider which machine learning model to use. At the time of this competition, I had read a lot about gradient boosting and the great successes it’s had in Kaggle Competitions, so I thought, let’s try It out with Xgboost!
I began by creating a function that performed K-Fold cross-validation for my Xgboost model. Since my dataset was relatively small (~600 data points), I explored the hyperparameter space using a Bayesian optimization Python package. The following is the code I used to set up the Bayesian optimization. The pbounds variable represents the parameter bounds of the hyper-parameter being investigated in the keys, and the tuple represents the range that can be explored by the Bayesian optimizer.

The Bayesian optimizer then performs optimizations and outputs the model performance for each model it creates. Below is a snippet of the Bayesian output in the Python console with Jupyter notebooks.
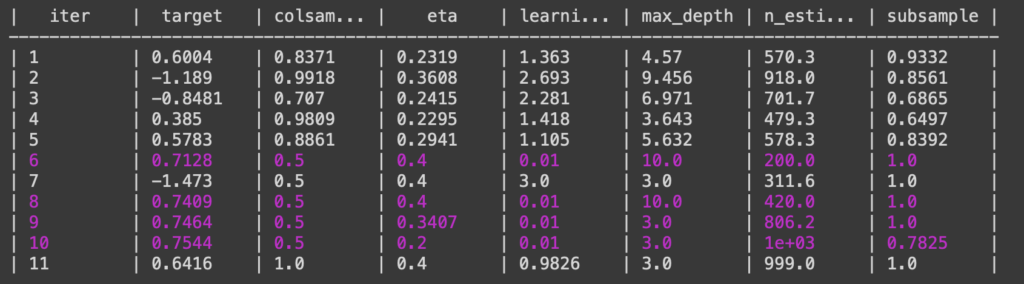
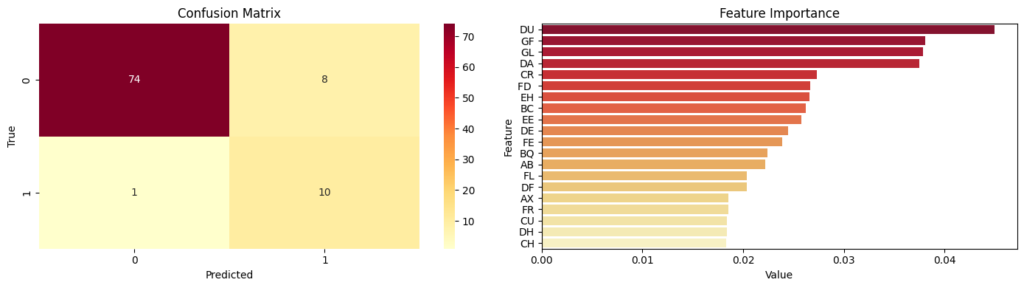
After the Bayesian optimizer runs for a certain amount of iterations, the best parameters are taken from the Bayesian optimizer and used to train a model. After the model was trained, a confusion matrix was created.
Final Competition Outcome
At the final competition evaluation, the hosts of the competition reevaluated everyone’s model based on a hold-out test set that was not included in the submission scores; the hold-out test set consisted of 400 test data points. When the final submission went through, I placed 4045/6431. I wouldn’t say I’m proud of this placement, but it did allow me to understand why my models weren’t performing as well as expected. I’d say that the main conclusion I came to from this was that I was overfitting my training data. Gradient boosting algorithms can easily do this, especially as the number of boosting rounds increases. Section 2 briefly explores alternative approaches to increase performance, but the Kaggle environment became problematic for notebook submissions to this competition.
Section 2: Alternative Approaches
After the competition closed, I decided to explore to explore two different avenues.
- Different model algorithms
- Different model embeddings
Model Algorithms
Sadly, when exploring this avenue, I ran into multiple problems with Kaggle’s environment since the competition submission was a Jupyter Notebook. I incorporated the following algorithms, but they didn’t submit within Kaggle’s competition.
- Random Forest Classifier
- Support Vector Machine Classifier
- MLPClassifier
I pinned the import of these packages from sci-kit learn as the issue, despite the notebook running when I saved it. So sadly, I reverted to the original Xgboost model, but this time I changed the hyperparameter windows. Originally I set a range of 200 to 1000 for n_estimators. The following are the new settings I used for hyperparameter optimization; n_estimators, learning rate, and max depth were all changed to smaller ranges.
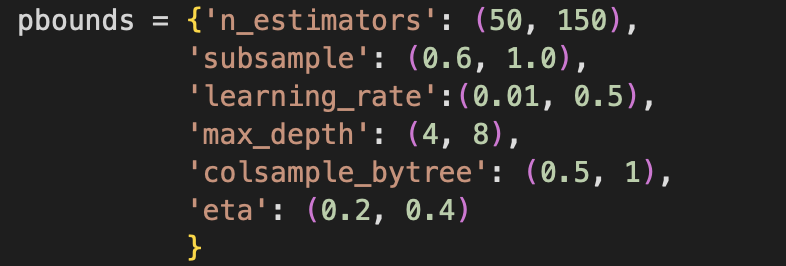
Submitting the new optimizations with a smaller range for some parameters did not make the performance go up. So I decided to explore more regularization parameters and added reg_lambda and reg_alpha to the hyperparameter optimization, which represents l2 norm regularization and the l1 norm regularization respectively. From a quick exploration of these parameters using Bayesian optimization, the performance did not increase. Again, I think this is pointing out that using a different model might be a wise decision.
Model Embeddings
I wanted to pursue the following embeddings to see how they would perform, but it became obvious that certain package imports were problematic with Kaggle’s environments.
- TruncatedSVD
- PCA
- UMAP
Conclusion
My model workflow would’ve benefitted from exploring different models, embedding types, and levels of regularization to see if I could produce a better model. Ultimately, this competition gave me a glimpse of what it may be like to work with patient data.